1. Introduction
2. Materials and Methods
Experimental organism
Light wavelength and photoperiod conditions
Total RNA extraction and cDNA synthesis
Real-time Polymerase Chain Reaction (real-time PCR)
In situ hybridization
Statistical analysis
3. Results
Per mRNA expression
Cry mRNA expressions
MT-R mRNA expressions
Confirmation of MT-R mRNA expression site
4. Discussion
1. Introduction
Light is an external environmental factor that influences the circadian rhythm of aquatic organisms, impacting their behavior, reproduction, and various biological responses (Pierce et al. 2008). All organisms exhibit different circadian rhythms in response to exposure to light. Circadian rhythms can be altered by seasonal changes, and artificial light not only modifies the circadian cycle but also alters the spectral composition of the light to which organisms are exposed, thereby affecting the fundamental 24 h circadian rhythm of aquatic organisms (Jin et al. 2009).
Circadian rhythms are internally regulated by clock genes and are often externally influenced by light, food, and temperature (Ripperger et al. 2011; Mata-Sotres et al. 2016). The prominent clock genes, period (Per) and cryptochrome (Cry), are regulated by light and enable organisms to perceive a 24 h cycle (Kim et al. 2018). Per forms a heterodimer with timeless genes and acts as a negative feedback loop that controls transcriptional activation and repression of clock genes (Landskron et al. 2009; Kim et al. 2012). Cry is rapidly induced in response to light stimulation by causing structural changes and autophosphorylation of Timeless through a direct interaction (Besharse et al. 2004). In addition to these clock genes, melatonin regulate the circadian rhythm in organisms. In invertebrates, melatonin is primarily expressed in the cerebral ganglia and retina (Gao et al. 2021). It is synthesized by arylalkylamine-N-acetyltransferase, which is inhibited by light, from the neurotransmitter 5-hydroxytryptamine and is released into the hemolymph at night, resulting in higher melatonin levels during the night than in the daytime (Iuvone et al. 2005). Melatonin plays a role in neuroendocrine transmission by regulating circadian rhythms and participates in immune regulation and antioxidant defense mechanisms (Petrovsky 2001; Maciel et al. 2010).
Although extensive research has been conducted on light-regulated clock genes and melatonin in terrestrial organisms (Terazono et al. 2003; Johnston et al. 2006), studies focusing on the physiological aspects of light responses in aquatic organisms, particularly invertebrates, remain relatively limited. Most existing research in this field has been conducted on fish species (Choi et al. 2019; Ma et al. 2023). Particularly, few studies on light wavelength-related physiology have focused on gastropods (invertebrates), such as abalones (Schnytzer et al. 2018), and studies on the expression characteristics of circadian rhythm-related genes are sparse (Gao et al. 2021; Botté et al. 2023).
Previous studies on nocturnal species, including abalones, have reported typical nocturnal behavioral patterns. They inhabit rocky reefs, hide in crevices during the day, and emerge to feed and engage in activities at night (Gao et al. 2021). Regarding nocturnal abalones, research conducted under controlled conditions showed that their survival and feeding rates were higher under constant darkness (DD), where 24 h darkness was maintained, compared to conditions with a natural light-dark cycle of 12 h light and 12 h darkness (12L:12D) (García-Esquivel et al. 2007; Gorrostieta-Hurtado et al. 2009).
Abalones, predominantly found in shallow coastal areas, are generally more light-sensitive than other fish species (Gao et al. 2022). Species inhabiting shallow depths are more responsive to long-wavelength light than those inhabiting deeper waters, and their spectral sensitivity peaks vary according to habitat depth, aligning with the spectral composition of their respective habitats (Gao et al. 2022). A study exposing abalones to various light wavelengths revealed that abalones exhibit photopositive behavior in the long-wavelength range (red and orange wavelengths), while avoidance behavior is observed in the short-wavelength range (blue and green wavelengths) (Gao et al. 2016). Furthermore, compared to the red and orange long-wavelength ranges, abalones exhibit significantly lower growth rates, survival rates, and feed conversion efficiencies in the blue and green short-wavelength ranges (Xiaolong et al. 2016).
These findings suggest that organisms perceive wavelengths in the long wavelength range, such as red light, as darker environmental conditions. This phenomenon indicates a close association between the perception of light wavelengths and various physiological responses related to circadian rhythms in abalones.
Therefore, this study aimed to investigate the effects of changes in the light-dark cycle and specific wavelengths of light on the circadian rhythms of abalones. Specifically, we examined changes in Per, Cry, and melatonin receptor (MT-R) mRNA expressions in abalones exposed to two light cycle conditions (12L:12D and DD) and red LED light. By examining the temporal expression patterns of these genes, particularly focusing on red wavelength light, which falls within the long wavelength range, we aimed to elucidate their impact on the circadian rhythm of abalones.
2. Materials and Methods
Experimental organism
Disk abalones (Haliotis discus hannai) (average length 6.35 ± 0.52 cm, average weight 42.07 ± 6.14 g) were purchased at an abalone farm (Wando, Jeollanam-do, Korea) and transferred to the laboratory for stability at laboratory tank conditions (salinity 34±0.7 psu, temperature 20.2±1.2°C, light cycle 12L:12D). The experiment was conducted over 24 h, and each experiment was conducted three times. During the adaptation period for two weeks, sufficient food (raw seaweed) was supplied once daily, and feeding was stopped 24 h before commencing the experiment. No organism died during the acclimatization and experimental periods.
Light wavelength and photoperiod conditions
During the experiment, the water quality conditions were maintained at 22°C, pH 8.0, and 35 psu under white fluorescent light (07:00 on, 19:00 off) and a thermostat (JS-WBP-170RP; Johnsam Co., Seoul, Korea) was used. The photoperiod group was illuminated with a white fluorescent bulb, while the red LED light-irradiated group received light from a red LED (Total LED Co., Seoul, Korea). Before starting the experiment, the experimental organisms were divided into photoperiod and LED groups (Fig. 1). The photoperiod group was exposed to two photoperiods (LD, 12L:12D; DD, constant darkness), and the LED group was exposed to a red LED (wavelength, 630 nm; 12L:12D). The experimental Zeitgeber time (ZT; 12L:12D) was determined based on natural light conditions. LD and LED groups lit at 07:00 and went off at 19:00 at an intensity of 0.5 W/m2 at the bottom of the water tank. Sampling was performed six times daily [ZT2 (09:00), ZT6 (13:00), ZT10 (17:00), ZT14 (21:00), ZT18 (01:00), and ZT22 (05:00) h]. A spectrometer (MR-16; Rainbow Light Technology, Pingzhen, Taoyuan, Taiwan) and an optical radiometer (HD 2102.1; Delta OMH Co., Caselle, Padova, Italy) were used. Retinal and hepatopancreatic tissues were collected and stored at -80°C until analysis.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from each retina and hepatopancreas sample using TRI Reagent® (TR 118; Molecular Research Center Inc., Cincinnati, OH, USA) according to the manufacturer’s instructions. Two micrograms of total RNA were reverse-transcribed in a total reaction volume of 20 μL using an oligo-(dT)15 anchor and M-MLV reverse transcriptase (M1701; Promega, Madison, WI, USA) according to the manufacturer’s protocol. The cDNA product was diluted and stored at 4°C for use in Reverse Transcription polymerase chain reaction (PCR) and real-time PCR analyses.
Real-time Polymerase Chain Reaction (real-time PCR)
Real-time PCR was conducted to determine the relative expression levels of Per and Cry associated with circadian rhythm genes using total RNA extracted from the abalone retina. Relative expression levels of MT-R were examined in the retina and hepatopancreatic tissues. Primers were designed using known disk abalone sequences (Table 1). PCR amplification was performed using a Bio-Rad iCycler iQ multicolor real-time PCR detection system (1708740; Bio-Rad Laboratories Inc., Hercules, CA, USA) and the iQ™ SYBR® green supermix (1708880; Bio-Rad Laboratories Inc., Hercules, CA, USA), following the manufacturer’s instructions. As a control, the β-actin gene was amplified from each sample. All data were expressed as the difference from the corresponding calculated β-actin threshold cycle (Ct) levels. The Ct values of the PCR products formed the basis for all analyses. These were defined as PCR cycles in which the fluorescence signal crossed a threshold during the exponential phase of the amplification curve. The calibrated ΔCt value (ΔΔCt) per sample and the internal control (β-actin) were calculated as follows: [ΔΔCt = 2- (ΔCt sample – ΔCt internal control)]. After the PCRs, real-time PCR data from three replicate samples were analyzed using the Bio-Rad system to estimate the transcript copy numbers for each sample.
In situ hybridization
The MT-R sequence, which was used as an in situ hybridization probe, was initially designed (Table 1) and then amplified using PCR. Subsequently, the plasmid was inserted into the pGEM-T Easy Vector (A137A; Promega, Madison, WI, USA). After confirming the antisense sequence via sequencing, the plasmid DNA underwent further amplification using PCR, employing antisense, sense, and T7 primers (5ʹ-TAA TAC GAC TCA CTA TAG GG-3ʹ). DIG-labeled RNA probes were synthesized using a DIG RNA labeling mix (Merck, Darmstadt, Germany), with PCR products obtained from the antisense primers and T7 RNA polymerase (Merck, Darmstadt, Germany) serving as antisense labeling probes.
Table 1.
Primers used for qPCR amplification and in situ hybridization
| Genes (Accession no.) | Primer | DNA sequences | E (%) |
| For qPCR | |||
| Per (MN547619) | Forward | 5ʹ–CGG TCC TGT ACC AGA TCC AC–3ʹ | 102.77 |
| Reverse | 5ʹ–CTG TAC CTC GAT CCA CTC GC–3ʹ | ||
| Cry (MW481327) | Forward | 5ʹ–GCT GCY ACA AAC AAC CCG TG–3ʹ | 98.48 |
| Reverse | 5ʹ–CAT GAT GGC GTC GAT CCA TG–3ʹ | ||
| MT-R (MW861452) | Forward | 5ʹ–GTG TCA TCA TCG GAA CCA TC–3ʹ | 101.14 |
| Reverse | 5ʹ–GTC AAG GTG ATA GTC TGG AC–3ʹ | ||
| β-actin (MW387000) | Forward | 5ʹ–GAT AGT GCG AGA CAT CAA GG–3ʹ | 95.38 |
| Reverse | 5ʹ–CGA TAG TTA TCA CAG ACC CG–3ʹ | ||
| For in situ hybridization | |||
| MT-R (MW861452) | Forward | 5ʹ–ATA TGT ACG TCA CGA CCT TG–3ʹ | |
| Reverse | 5ʹ–CAT GCG CAG ATA ACA GAT TC–3ʹ |
To prepare frozen section tissue slides, we collected hepatopancreatic tissues from disk abalones across all experimental groups, with some groups sampled at 6 h (in the LD and RD groups) and others at 18 h (in the RD and DD groups). The collected tissues were then fixed overnight at 4°C in a 4% paraformaldehyde solution. After fixation, the samples were washed with phosphate-buffered saline (PBS) and stored in a 30% sucrose solution to prevent the formation of ice crystals. Next, the sections were subjected to hybridization using a hybridization buffer comprising deionized formamide, 20× saline sodium citrate, 0.1% Tween-20, 1 M citric acid, and yeast tRNA (in a 1:100 ratio with the hybridization buffer). The RNA probe was included simultaneously, and hybridization was carried out for 18 h at 65°C.
To detect the hybridization signal, the tissue sections were first exposed to a blocking solution containing calf serum in PBS/Tween (PBST) for 1 h at 20°C. Subsequently, the sections were incubated at 4°C with an anti-digoxigenin antibody conjugated to alkaline phosphatase (at a 1:2000 dilution in the blocking solution; Roche, Basel, Switzerland). After multiple PBST washing steps, the sections were rinsed in alkaline Tris buffer consisting of 1 M Tris at pH 9.5, 1 M MgCl2, 5 M NaCl, and 10% Tween-20. Color development was achieved using a labeling mix of alkaline Tris buffer, nitro blue tetrazolium, and 5-bromo-4-chloro-3-indolyl phosphate disodium salt. Finally, the sections were washed with PBST, mounted with Aqua Polymount (Polysciences, Warrington, PA, USA), and observed under a stereomicroscope (Nikon Eclipse CI, Tokyo, Japan) for image capture.
Statistical analysis
All data were analyzed using SPSS v. 24.0 (IBM Corp., Armonk, NY, USA). A two-way ANOVA and Tukey’s post hoc test were used to compare differences in the data (P < 0.05). Values are expressed as mean±standard deviation (SD).
3. Results
Per mRNA expression
Per mRNA expression peaked at 2 h of exposure, regardless of the light source conditions, and Per expression significantly decreased over time (Fig. 2) (P < 0.05). The expression decreased in the RD group compared to the LD group, and the expression level decreased the most in the DD group. From 14 to 22 h, there was no significant difference in Per mRNA expression between the RD and DD groups (P > 0.05).

Fig. 2.
Expression of period (Per) mRNA in the retina of disk abalone exposed to different wavelength and light cycle treatments. The letters indicate significant difference according to the same light treatments at the different zeitgeber time (P < 0.05). The numbers indicate significant differences according to the different light treatments at the same zeitgeber time (P < 0.05). All values are expressed as means ± standard deviation (n = 5)
Cry mRNA expressions
The Cry mRNA expression peaked at 6 h and increased after 2 h of exposure (Fig. 3). Cry mRNA expression decreased significantly over time (P < 0.05) and was maintained from 18 h. Cry mRNA expression was significantly higher in the LD group than in the RD group, and the expression level decreased the most in the DD group (P < 0.05). From 14 h to 22 h, there was no significant difference in mRNA expression levels between the RD and DD groups.

Fig. 3.
Expression of cryptochrome (Cry) mRNA in the retina of disk abalone exposed to different wavelength and light cycle treatments. The letters indicate significant difference according to the same light treatments at the different zeitgeber time (P < 0.05). The numbers indicate significant differences according to the different light treatments at the same zeitgeber time (P < 0.05). All values are expressed as means ± standard deviation (n = 5)
MT-R mRNA expressions
Except for the RD group, retinal MT-R mRNA expression was maintained at a constant level from 2 h to 10 h of exposure and then increased over time from 14 h (Fig. 4a). Thereafter, a peak was observed at 18 h, and MT-R mRNA expression significantly decreased at 22 h (P < 0.05). MT-R mRNA expression was significantly higher in the DD group than in the RD group, and the expression level decreased the most in the LD group (P < 0.05). From 10–22 h, there was no significant difference in mRNA expression levels between the RD and DD groups.
In the hepatopancreas, MT-R mRNA expression peaked at 18 h and decreased significantly at 22 h (Fig. 4b) (P < 0.05). MT-R mRNA expression was significantly higher in the DD group than in the LD group in 14–22 h (P < 0.05). However, there were no significant differences in the mRNA expression levels between the LD and RD groups from 6–10 h and between the RD and DD groups from 14–22 h.
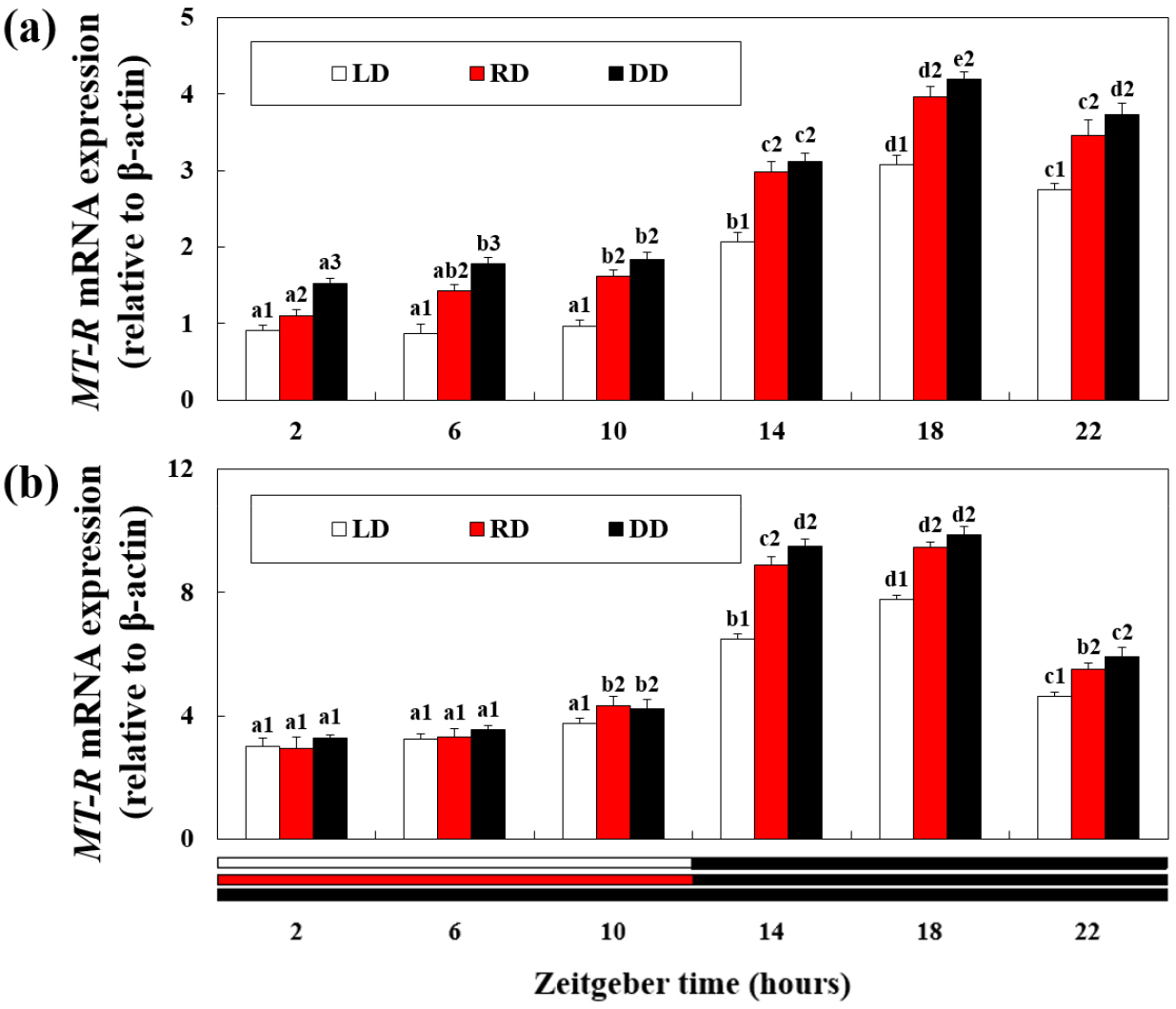
Fig. 4.
Expression of melatonin receptor (MT-R) mRNA in the (a) retina and (b) hepatopancreas of disk abalone exposed to different wavelength and light cycle treatments. The letters indicate significant difference according to the same light treatments at the different zeitgeber time (P < 0.05). The numbers indicate significant differences according to the different light treatments at the same zeitgeber time (P < 0.05). All values are expressed as means ± standard deviation (n = 5)
Confirmation of MT-R mRNA expression site
To confirm MT-R mRNA expression in the hepatopancreas, in situ hybridization was performed using an MT-R mRNA probe (Fig. 5). MT-R mRNA signal intensity was observed in the cytoplasm of the hepatopancreatic tissue, consistent with MT-R mRNA expression. When comparing the expression levels between the LD and RD groups at 6 h when light was provided, the MT-R probe signal was notably higher in the RD group than in the LD group. Conversely, when comparing expression levels between the RD and DD groups during the dark period of 18 h, a significantly stronger signal was observed in the DD group.
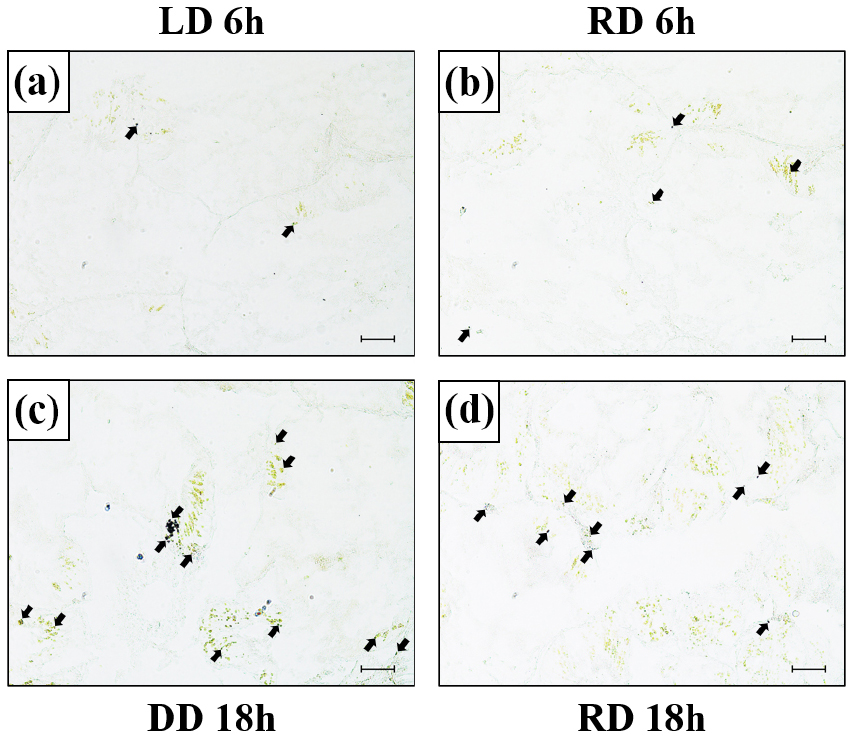
Fig. 5.
Expression of the melatonin receptor (MT-R) mRNA in the hepatopancreas of disk abalone detected by in situ hybridization (a–d). (a) LD group at 6 h, (b) RD group at 6 h, (c) DD group at 18 h, and (d) RD group at 18 h. Dark areas (black arrow) indicate the mRNA expression of MT-R in the hepatopancreas. Scale bar = 50 μm
4. Discussion
The circadian rhythm is a key factor influencing various physiological functions in organisms, including growth, maturation, and behavior (Patke et al. 2020). Changes in the external natural environment, such as alternating light conditions between day and night, can lead to physiological changes in organisms.
In our study, exposure to decreased light intensity during the transition from the light-to-dark period led to a significant decrease in the expression levels of Per and Cry mRNA in abalones. Kong et al. (2023), who investigated the expression of Per and Cry for 24 h in razor clams (Sinonovacula constricta), a nocturnal shellfish similar to abalones, reported that the expression levels of Per and Cry significantly increased during the light period and started decreasing at the onset of darkness. This indicates a rhythmic expression pattern influenced by the light response. The increased expression of Per and Cry observed in this study also indicates light-responsive regulation, suggesting that the presence or absence of light influences the expression of circadian clock genes. In contrast, the MT-R, known to be expressed at night, showed a significant increase in expression during the transition from day to night in our study. In situ hybridization results confirmed a high level of expression at night (18 h). This finding is consistent with the results of Balzer et al. (1997), who conducted a study on crayfish (Procambarus clarkii), an aquatic invertebrate species, and reported increased MT-R expression during the dark period. These results collectively suggest that a decrease in light intensity leads to increased melatonin synthesis under dark conditions, resulting in the upregulation of MT-R expression. Increasing melatonin levels enhances antioxidant capabilities and reduces oxidative damage by eliminating reactive oxygen species in the body (Zhang et al. 2019). Additionally, it promotes the expression of immune proteins, thereby enhancing the organism’s immune system (Huang et al. 2020).
Since the abalones in this study were exposed to DD conditions without light, owing to the expression characteristics of each gene, it was expected that MT-R would consistently show an extremely high expression level and Per and Cry would consistently show an extremely low expression level for 24 h. Despite the absence of light, the expression levels of MT-R, Per, and Cry in DD-exposed abalones were similar to those observed in abalones exposed to LD conditions. It is worth noting that Per and Cry, which are the key regulators of circadian rhythms, are typically regulated by external factors such as light. However, even in the absence of light, these genes are regulated to some extent by internal negative feedback mechanisms, allowing for a certain degree of rhythmicity following a 24 h cycle (Fonken and Nelson 2014). The presence of circadian rhythms in clock genes has been reported in bivalve species such as oysters (Crassostrea gigas) (Perrigault and Tran 2017) and mussels (Mytilus edulis) (Chapman et al. 2020). However, rhythmicity has not been demonstrated in marine gastropods, such as sea snails (Bulla gouldiana) (Constance et al. 2002) and limpets (Cellana rota) (Schnytzer et al. 2018).
Therefore, based on the observed consistent gene expression patterns, abalones may have their intrinsic circadian rhythms, distinguishing them from other marine gastropods, even under conditions without light (DD). However, further research on the molecular mechanisms and functions of clock genes is needed to clarify and better understand circadian rhythms in invertebrates.
Another noteworthy finding in this study was the decrease in Per and Cry expression and the increase in MT-R expression in the RD experimental group (red LED exposure group) compared to the control group (LD group). This pattern was partially similar to the expression trends observed in the DD experimental group, where darkness was maintained. Additionally, there was no significant difference in the expression of clock genes between the RD and DD experimental groups, starting from the onset of darkness at 14 h. Considering these two results, it can be inferred that abalones perceive red wavelengths as darkness, leading to similar circadian rhythms under RD and DD conditions. A study by Jang et al. (2017) on mud shrimps (Upogebia major) also reported that exposure to a 650 nm red wavelength did not induce light-stimulated responses similar to the DD condition. Based on the similarity between the circadian rhythms observed in the RD and DD conditions in this study, it is expected that effects such as growth and maturation promotion reported under DD conditions (Xiaolong et al. 2018) may also occur under RD conditions.
In summary, in this study, we investigated the effects of two light cycle conditions (12L:12D, DD) and the red wavelength in the light environment on the expression pattern of clock genes in abalones. We found that red LED light induced gene expression patterns closer to the 24 h memorization condition. These results suggest that long- wavelength red light can alter the circadian rhythm in abalones, in contrast to the effects of general light exposure. The findings of this study provide valuable scientific information on the circadian rhythm of abalones (marine gastropods), which is less understood than that of fish, and contribute to our understanding of the physiological responses of nocturnal abalones to changes in light wavelengths.





