1. Introduction
2. Materials and Methods
Collection of samples and culture condition
Morphological identification
Molecular identification
3. Results and Discussion
1. Introduction
The genus Surirella (Bacillariophyta, Bacillariophyceae, Surirellales) was first described in 1828 by Turpin (Krammer and Lange-Bertalot 1999). This genus is one of the very old and taxonomically rich genera belonging to family Surirellaceae. Members of this genus are widely distributed worldwide in all benthic environments of freshwater, brackish, and seawater, although they generally have high richness and diversity in tropical environments (O'Sullivan and Reynolds 2004; Bramburger et al. 2006).
Van Landingham (1978) has reported 621 Surirella species. A total of 1,358 Surirella species are currently described in Algaebase, including 579 infraspecific names. Of these, 262 species have been transferred to other genera (Genus Iconella, Campylodiscus, Cymatopleura, etc.) or considered synonyms. So far, only 243 scientific names have been recognized based on literature (Guiry and Guiry 2011). Many species of genus Surirella have been reported only once or several times in type locality (Veselá et al. 2013), which might be one of the reasons why some 800 species are still not clearly recognized despite their high diversity. In addition, genus Surirella was traditionally divided into three subgroups (Fastuosae, Pinnatae, Robustae) based on differences in keel morphology and associated support structures (Krammer and Lange-Bertalot 1999). However, Ruck and Kociolek (2004) have reported that the genus Surirella is not a monophyletic group. Ruck et al. (2016) have suggested that existing groups in Surirellaceae should be newly defined to identify them. Such taxonomic complexity can make a clear understanding of this genus even more difficult. Currently, only 25 species in genus Surirella have been reported in Korea, excluding synonyms (NIBR 2020; MABIK 2021).
In this study, we isolated and identified Surirella atomus from a mudflat of Hampyeong Bay on the west coast of Korea. We report the first record of this species in Korea and information on its morphology and molecular features.
2. Materials and Methods
Collection of samples and culture condition
Samples were collected from intertidal mudflat in Hampyeong Bay (35º03'43.12''N, 126º24'53.68''E), west coast of South Korea in July 2018 (Fig. 1).
Samples were examined under an Eclipse Ti-U inverted microscope (Nikon, Tokyo, Japan) and single cells were isolated and transferred to the cell culture flask (SPL Life Sciences, Pocheon, Korea) with f/2 medium (with silicate, Sigma Aldrich Co., St. Louis, MO, USA) by the capillary method. The antibiotics (penicillin-streptomycin-neomycin) mixture (Sigma Aldrich Co., St. Louis, MO, USA) was added to culture media for inhibition of bacterial growth. Strains were incubated at 10°C at a 14:10 h light dark cycle. Illumination was provided by a fluorescent lamp with an irradiance of 100 μmol photons m-2s-1. The strains were transferred to fresh medium at four-week intervals.
Morphological identification
Strains were cleaned with acid to clean for frustules (Trobajo and Mann 2019). For light microscope examination, permanent slides were made using Mountmedia (Wako Pure Chemical Industries, Osaka, Japan) and examined using Eclipse Ni stereoscopic microscope (Nikon, Tokyo, Japan). For scanning electron microscope examination, strains fixed with Lugol’s solution were filtered in PC membrane (25 mm; pore size 1 or 2 μm) and then washed with distilled water. The membranes were dehydrated in a graded ethanol series (10~100%) and dried using tetramethylsilane (Sigma Aldrich Co., St. Louis, MO, USA). Finally, the membranes were mounted on stubs and sputter-coated with gold. Observations were made with Hitachi S-4300 scanning electron microscope (Hitachi, Tokyo, Japan) and Sigma 500-VP field emission scanning electron microscope (Ziess, Göttingen, Germany). Slides (MABIK DI00042892) and SEM stub (MABIK DI00042893) used this study were deposited at the National Marine Biodiversity Institute of Korea (MABIK).
Molecular identification
The cultured strain was harvested by centrifugation at 13,000 rpm for 1 min and then the pellets were mixed in 1 ml of sterilized STE buffer solution. Genomic DNAs from each samples were extracted using DNeasy PowerSoil Pro Kit (Qiagen Inc., Hilden, Germany) following the manufacturer’s protocol. The DNA of strain was amplified at two genetic regions: large subunit of the ribulose- bisphosphate carboxylase gene (rbcL) and apocytochrome b gene (cob). The rbcL region was amplified using DPrbcL1 and DPrbcL7 primers (Daugbjerg and Andersen 1997), and the cob region was amplified using cobF124 (5′-gcttggagttttggdtcttydgc-3′) and cobR847 (5′-ggyaaaaartaycaytcmggbac-3′) primers (unpublished). The overall experiment for PCR were performed following the protocol described in An et al. (2017). PCR products were purified using ExoSAP-IT Express PCR Product Cleanup Reagent (Thermo Fisher Scientific, MA, USA) and sequenced by the Automatic DNA sequencher, ABI Prism 377, Applied Biosystems (Macrogen, Seoul, South Korea).
Sequence was assembled and edited using Geneious R7 v.7.1.2 (Biomatters Ltd., Auckland, New Zealand). Sequences for each of the two regions were aligned both individually using ARB program package (Ludwig et al. 2004). A data set of concatenated rbcL and cob sequence data was constructed, including 35 surirellaceae sequences from GenBank (Table 1).
Table 1.
A list of species, specimens and GenBank accession numbers of sequences used in this study
Phylogentic tree was constructed using maximum likelihood (ML) and Bayesian phylogenetic inference (BI) with Randomized Axelerated Maximum Likelihood v.8.2.1 (RAxML) (Stamatakis 2014) and MrBayes version 3.2.7 (Ronquist and Huelsenbeck 2003). The ML analysis was performed under default settings. Bayesian phylogenetic inference (BI) was performed following methods described in Ruck et al. (2016). Thalassiophysa hyalina (Grunow) Gran served as the outgroup.
3. Results and Discussion
Genus Surirella Turpin 1828
Type species:Surirella striatula Turpin 1828
Surirella atomus Hustedt 1955
Type locality: marine littoral, Beaufort, North Carolina (Hustedt 1955: 48)
Specimens examined: Specimens were obtained from mudflat in the Hampyeong Bay, Muan-gun, Jeollanam-do (35º03'43.12''N, 126º24'53.68''E), on the West Coast of Korea on July 19, 2018.
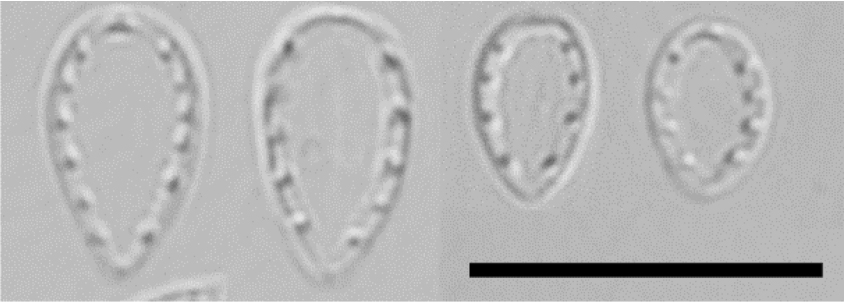
Observation: Frustules are spatulate in valve view and rectangular (or wedge-shaped) in girdle view. Heteropolar valve is broadly ovate with a broadly rounded head pole and cuneately rounded foot pole, 5–8 μm long, 3–4 μm broad (Fig. 2). The striae are numerous, radiate at apices, parallel in the middle, 65 in 10 μm and costae and central line are not visible in the LM. The fibulae are broad and short with 10–11 in 10 μm.
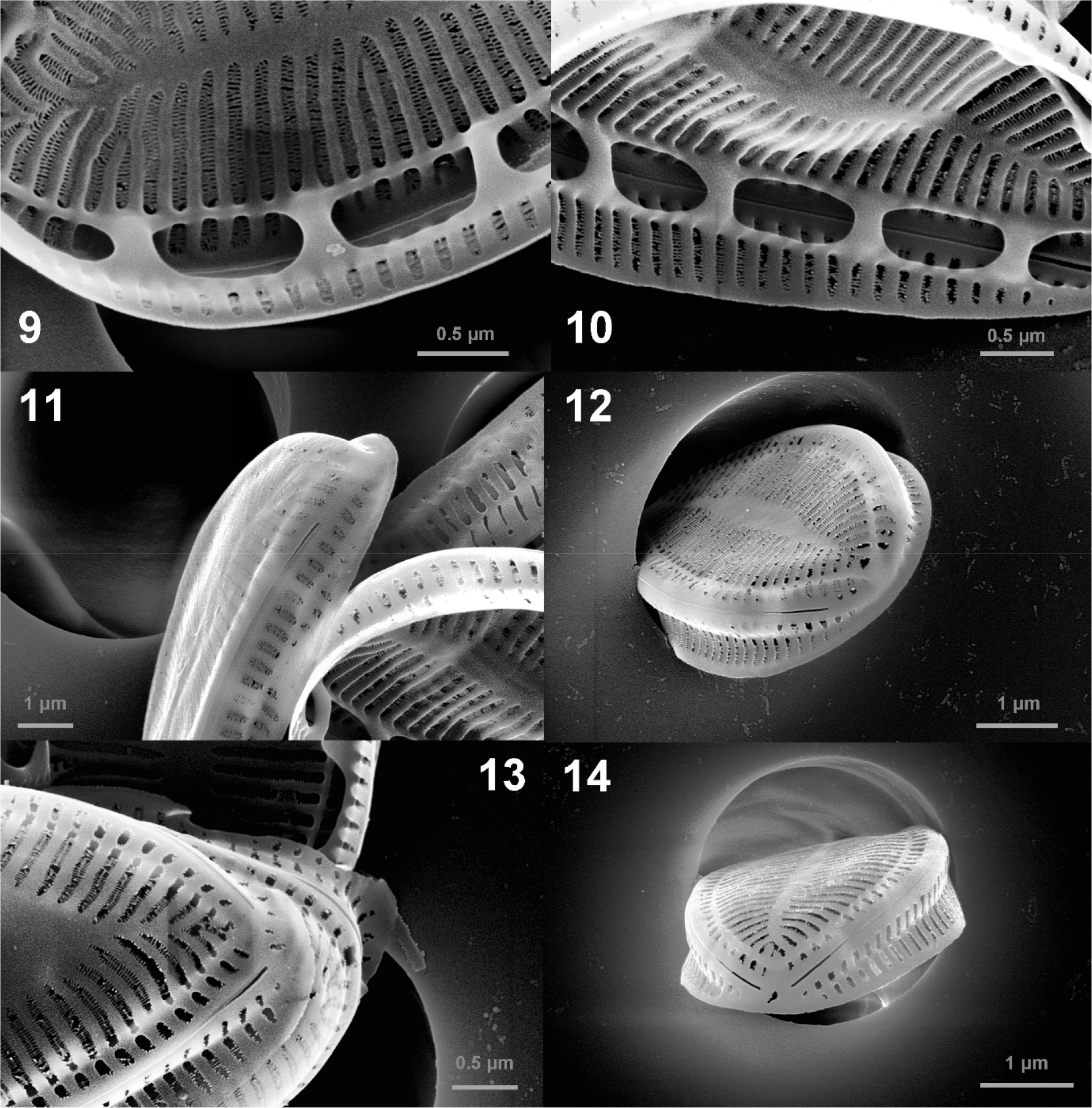
In SEM, valve face is flat, and there is no ornamentation such as spine, tubular process or siliceous tendrils (Figs. 3, 4, 5, 6, 7). The areolae are arranged in two rows throughout the valve and sometimes in three rows at the valve margin (Fig. 8). The areolae are oblong in both external and interior view (Figs. 8 and 9). The multiseriate striae run from the midline to the valve margin (Figs. 3 and 4). The fibulae are narrow (110–200 nm) and short (ca. 300 nm) (Figs. 9 and 10). The fenestrae (portulae) are oval i n shape, easily observed opening into the keel canal and fenestral bar is absent. The mantle is slightly concave without ornamentation. The mantle has areolae and striae of the same shape as the valve, and line up with those on the valve face (Fig. 11). Ridged raphe fissure is interrupted at both the head and the foot poles and raphe ends are simple with straight unexpanded endings internally and externally (Figs. 11, 12, 13, 14).
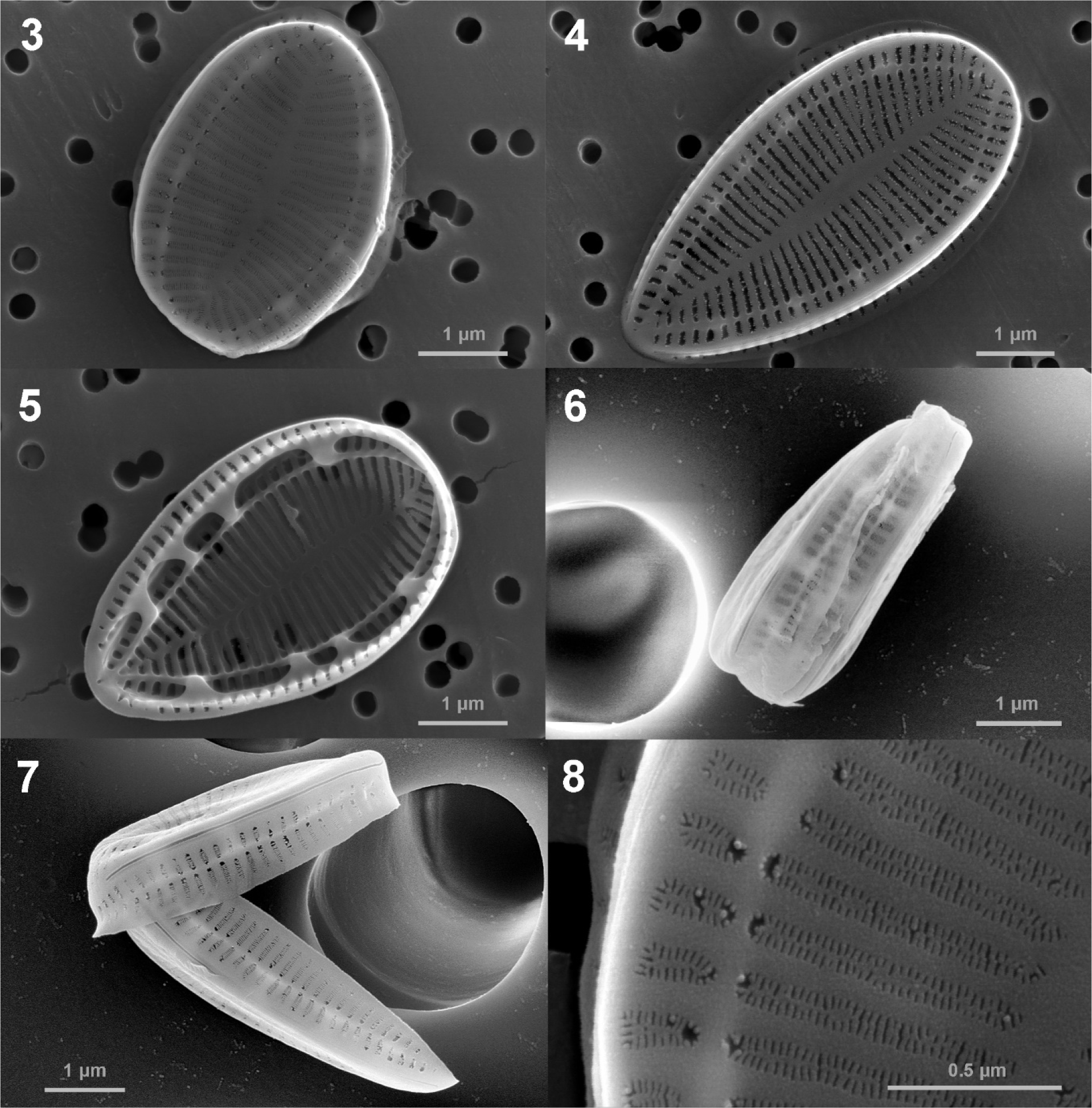
Fig. 3-8.
SEM micrographs of the Surirella atomus, external (Figs. 3–4, 6–8) and internal (Fig. 5) views Figs. 3 and 4. external valve views of whole valve; Fig. 5. internal views of whole valve; Figs. 6 and 7. external girdle views of whole valve; Fig. 8. Detail of valve showing the oblong areolae and multiseriate striae
Remark: Few literature has described morphological characteristics of this species. Surirella atomus is smaller than most other species of Surirella. The original description has described that this species is the smallest species of genus surirella observed (Hustedt 1955). Our specimens are shorter and narrower than the morphometrics given for this species in the original literature (about 12 μm long, 6 μm broad). However, other studies have also recorded smaller specimens of this species (English 2011; Morais et al. 2018). The length/width ratio and number of fibulae in 10 μm of the obtained strain support the original literature. In LM observation, these specimens are similar to S. stalagma and S. suecica. However, S. atomus is usually smaller than S. stalagma and S. suecica. S. stalagma has capitate footpole and more narrow headpole. S. suecica has broader and visible fibulae.
Molecular identification: In this study, partial gene sequences of the rbcL (1,433 bp) and cob (660 bp) of Surirella atomus were obtained for the first time. These sequences have been deposited in GenBank (MZ062029 and MZ090958). We compared our sequences with sequences in Genbank using BLASTn algorithm. In the case of the rbcL gene, S. atomus shared more than 97.1% sequence similarities with Petrodictyon gemma (KX120607). However, in the case of cob gene, this species shared a low sequence similarity (82.2%) with Stenopterobia pumila (JX032867).
Phylogenetic analysis was conducted to establish the relationship between S. atomus with other surirellaceae species (Fig. 15). S. atomus was weakly (bootstrap value = 59) supported as sister to Petrodictyon gemma. Ruck et al. (2016) have proposed a genus Petrodictyon + S. febigeri as a phylogenetically uncertain placement. Although further research on molecular phylogenetic analysis is needed, results of this study show that S. atomus is also in the same situation. However, considering the replacement criteria of genus in Surirellaceae based on morphological characteristics proposed by Jahn et al. (2017) and Ruck et al. (2016), it seems clear that this species belongs to genus Surirella (Table 2).
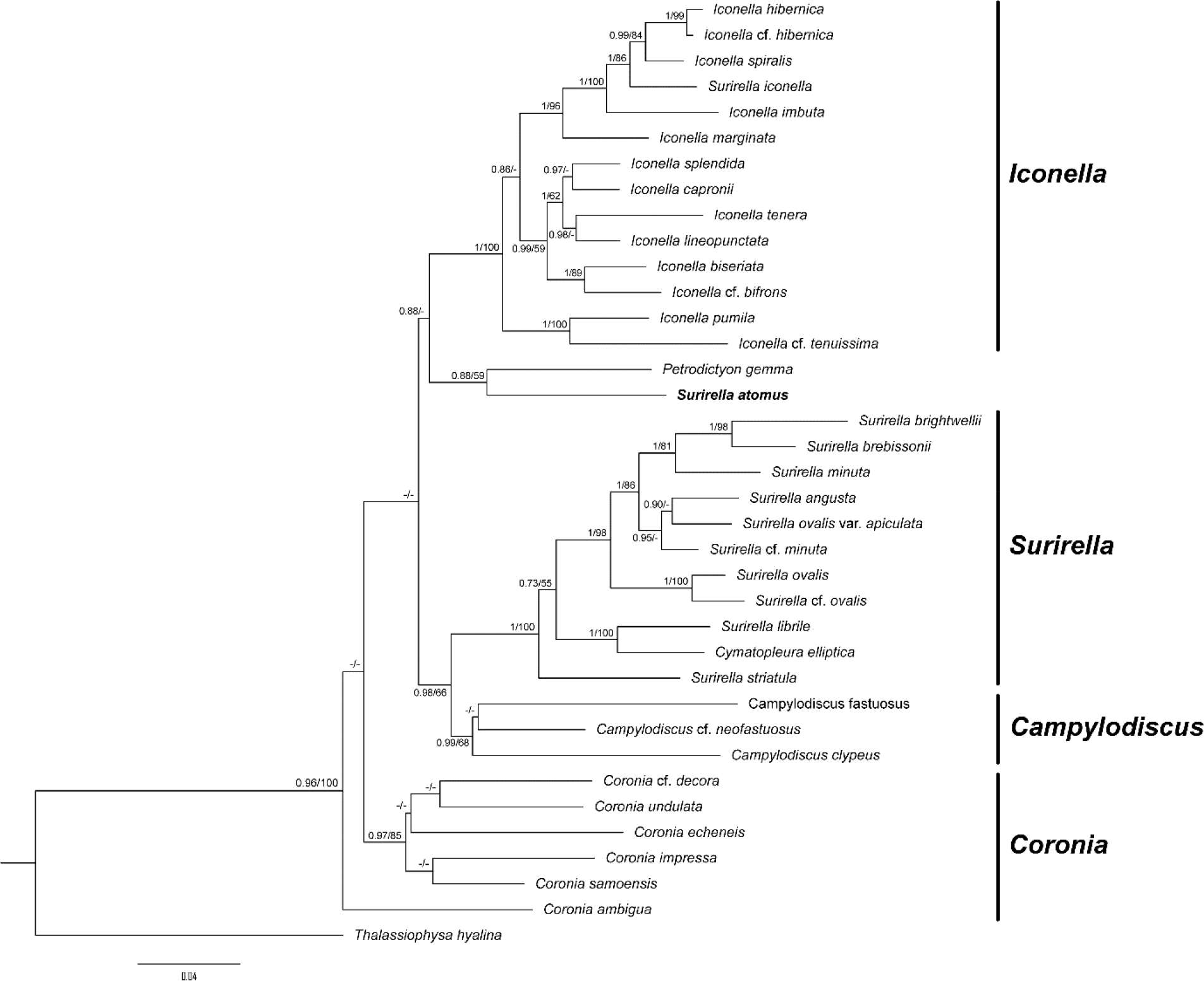
Fig. 15.
A phylogenetic tree obtained from ML and BI analysis based on the concatenated dataset showing the phylogenetic positions of Surirella atomus. The values on each node indicate Bayesian posterior probabilities and bootstrap, respectively. Values less than 50% ML bootstrap and 0.7 BI are indicated as ‘-’
Table 2.
Comparison of morphological characteristics of major genus within Surirellaceae and Suriella atomus (Jahn et al. 2017; Ruck et al. 2016)
Distribution and habitat: This species is known as a marine species. Bate et al. (2004) have reported the salinity range in which they appeared as 13–18 ppt, and Denys (1991) has classified this species as a mesohalobous euryhaline species. In this study, this species was founded in a tidal flat with a salinity of 28.5 PSU. Previous studies have confirmed that most of them are found in marine and brackish environment, including Alabama Coast, southeastern Louisiana and St. Louis Bay in USA (O'Neil 1982; Sullivan 1982; Maples 1983), Great Fish Estuary in South Africa (Bate et al. 2004), Gulf of Mexico (Krayesky et al. 2009), Tagus estuary in Portugal (Vieira et al. 2013), Jiaozhou Bay in China (Qi et al. 2016), Loire estuary in France (Barillé et al. 2017), Porto-Novo Lagoon in Benin (Akogbeto et al. 2018), Dalyan Beach, Turkey (Kaleli et al. 2020). However, a few of them have been found in fresh water, such as La Trobe River in Australia (Chessman 1985), Montana, South Dakota and Wyoming in USA (Bahls 2009) and Martinjärvi and Iso Kivijärvi in Finland (Nikolajev-Wikström 2015). Considering regions where it has been found, this species seems to be mainly distributed in the mid-latitude temperate region. Whether it is a marine species requires additional review.
According to the original description by Hustedt (1955), this species is a very rare species. However, this species has a relative frequency of more than 5%, and it has been reported as an indicator of eutrophication at Porto-Novo Lagoon, Benin (Akogbeto et al. 2018). In addition, this species showed a high relative frequency of over 2.5% in summer and autumn at La Trobe River, Australia (Chessman 1985).
This species was collected from sediment in mudflat in this study. Sullivan (1982) has reported that this species is only observed in sediments in a study of epiphytic diatoms communities on red algae. However, this species has been reported as an epibiont attached to various substrates, including pneumatophores of mangrove (Maples 1983), oysters (Barillé et al. 2017), and turtle shells (Kaleli et al. 2020). Thus, although this species is basically an epipelic diatom, it seems that it also prefers solid substrates.