1. Introduction
2. Materials and Methods
Sample collection and isolation
Morphological characterization
Phylogenetic characterization
3. Results and Discussion
Morphological characteristics
Phylogenetic analysis
1. Introduction
The genus Melosira Schmidt includes diatoms classified under the family Melosiraceae Kützing, and is a highly branched taxonomic group with wide global distribution (Round et al. 1990). Melosira species are well known for their ecological adaptability, as they can thrive in diverse environmental conditions, ranging from oligotrophic to eutrophic waters and from freshwater lakes to marine estuaries (Fischer 1986).
Members of the genus Melosira are characterized by their cylindrical or subspherical cell shape and often form chain-like colonies with mucilage pads and small irregular spines (Round et al. 1990). Two to three cells within a colony are typically connected by a cingula. The frustum exhibits equicircularity and radial symmetry. Valves may exhibit chambered structures and external characteristics such as small spines and granules, especially in regions where adjacent cells come into contact and establish connections. Furthermore, a circular flange, referred to as a corona or carina, may be observed at a specific distance from the center of the valve (Crawford 1975). Rimoportulae were distributed either individually or in clusters on the valve face and mantle. The loculi were organized in either a random pattern or in rows radiating outward from the center. The cingulum exhibited ligulate bands with small pores organized in longitudinal rows (Houk et al. 2007).
The genus Melosira was established by Agardh (1824), with Melosira nummuloides Agardh proposed as the type species. However, the first reported species of this genus was Conferva moniliformis Müller (= Melosira moniliformis Agardh), which was reported by Müller in 1783 (Houk et al. 2007). Hustedt (1930) considered the taxa Orthosira Thwaites and Paralia Heiberg to be synonymous with Melosira, leading to a significant increase in the number of species attributed to this genus (Houk et al. 2007). Hustedt’s classification was widely used in academia until the advent of electron microscopy (Werner 1977). Subsequently, technological advancements have resulted in the revision of the current classification system (Round et al. 1990), with the addition of numerous new species (Fernandes and Souza-Mosimann 2001; Kaczmarska and Jahn 2006; Van de Vijver and Crawford 2020; Yang et al. 2022). To date, 833 species have been documented within the genus Melosira, with 86 species, including subspecies, varieties, and formae, currently recognized taxonomically (Guiry and Guiry 2024). According to the latest reports, 11 species within the genus Melosira have been documented in South Korea, excluding synonymous species (NIBR 2022; MABIK 2023).
Melosira octogona Schmidt was initially documented by Schmidt (1893, pl. 182: figs. 19–21), and is considered to have a global distribution, with occurrences reported in Europe, the Americas, Asia, Africa, and Australia (Guiry and Guiry 2024). In South Korea, this species was first reported by Chung (1969) in the Han River estuary. Subsequent reports indicated its presence in the Geum River estuary (Cho et al. 1978) and the Yellow Sea (Cho et al. 1983). Although the global distribution of this species has been confirmed, knowledge of its ultrastructure and DNA sequences is limited. Consequently, in this study, M. octogona was isolated and identified from coastal waters off Jeju Island, South Korea. This study represents the first investigation of this species and provides insights into its ultrastructure and small subunit (SSU) sequence using scanning electron microscopy (SEM) and molecular analysis.
2. Materials and Methods
Sample collection and isolation
A sample was obtained from the coastal water at Ojo-ri, Seongsan-eup, Seogwipo-si, Jeju Island, South Korea (33° 27'3.7"N, 126°55'15.1"E), on May 25, 2022 (Fig. 1). The water depth within the survey area was notably shallow, measuring approximately 10 cm, and there was almost no water flow. The water temperature and salinity at the time of sampling were 29.6°C and 6.6 psu, respectively. The seawater sample was obtained from the surface layer using a plastic beaker, ensuring that sediment was not introduced. Subsequently, it was transferred to the laboratory in a 0.5-liter sterile water collection bottle. Cells were isolated using the capillary method under an Eclipse Ti-U inverted microscope (Nikon, Tokyo, Japan). The isolated cells were further transferred to a cell culture flask (SPL Life Sciences, Pocheon, South Korea) with F/2 medium supplemented with silicate (Sigma-Aldrich Co., St. Louis, USA) at a 10‰ salinity level. The culture strains were subsequently incubated under conditions of 20℃ temperature, a 14 h/10 h light/dark cycle, and an irradiance of 40 μmol m-2 s-1.
Morphological characterization
The cultured strains were examined using an AX10 light microscope equipped with an Imager A2 digital camera system (Zeiss, Göttingen, Germany). Examination was performed using a 100× Plan-Apochromat oil-immersion objective lens (1.30 N.A.). Examination using a SEM was performed according to the protocol described by An et al. (2022). In brief, the cultured strain was fixed in 5% Lugol’s solution, filtered through a polycarbonate membrane measuring 25 mm in diameter and with a 3 μm pore size (Advantec, Tokyo, Japan), and subsequently rinsed three times with sterile distilled water. The membrane was dehydrated using a graded series of ethanol solutions (10–100%). The samples were dried using tetramethylsilane (Sigma-Aldrich Co., St. Louis, USA). The membrane was affixed to a stub and coated with gold using an MC1000 ion sputterer (Hitachi, Tokyo, Japan). Specimens were examined using a high-resolution Zeiss Sigma 500 VP field-emission SEM (Carl Zeiss, Oberkochen, Germany).
Phylogenetic characterization
The culture strain was acquired through centrifugation at 2,500 rpm for 5 min, after which the supernatant was discarded. Genomic DNA was extracted using the DNeasy PowerSoil Pro Kit (Qiagen Inc., Hilden, Germany) following the manufacturer’s guidelines. PCR was performed according to the procedure described by An et al. (2022). The primer pairs Diatom9F (Lynch et al. 1997) and EukBR (Medlin et al. 1988) were used to amplify the 18S rDNA region. The PCR product was purified using the ExoSAP -IT Express PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and subsequently sequenced by Cosmo Genetech Inc. (Seoul, South Korea). The sequences were trimmed, assembled, and aligned using Geneious Prime version 2023.2.1. (Biomatters Ltd., Auckland, New Zealand). Phylogenetic trees were generated using the Randomized Axelerated Maximum Likelihood (RAxML) version 8.2.10 (Stamatakis 2014) and MrBayes version 3.2.7 (Ronquist and Huelsenbeck 2003), by employing the maximum likelihood (ML) and Bayesian phylogenetic inference (BI) methodologies, respectively. ML analysis was performed using the GTR GAMMA model with 1,000 bootstrap replicates while maintaining default values for all other settings. BI analysis was performed using the established methods detailed by An et al. (2022). A dataset comprising 18S rDNA sequences from 23 Melosirales species obtained from GenBank was compiled. The trees were visualized using Figtree version 1.4.4 and Adobe Illustrator version 28.1 (Adobe Systems, San José, USA).
3. Results and Discussion
Morphological characteristics
Cells were arranged in short chains, with frustules exhibiting an octagonal shape in girdle view because of straight, obliquely oriented valve face margins (Fig. 2a). Small, plate-like, golden-yellow plastids were observed in the peripheral cytoplasm (Fig. 2a). They were distinctly united into pairs by their cingula (Fig. 2a, b and d). The corona was well-developed in valve face (Fig. 2c). The valve diameters ranged from 10.4 to 11.1 μm (n = 10), while the height ranged from 9.9 to 10.6 μm.
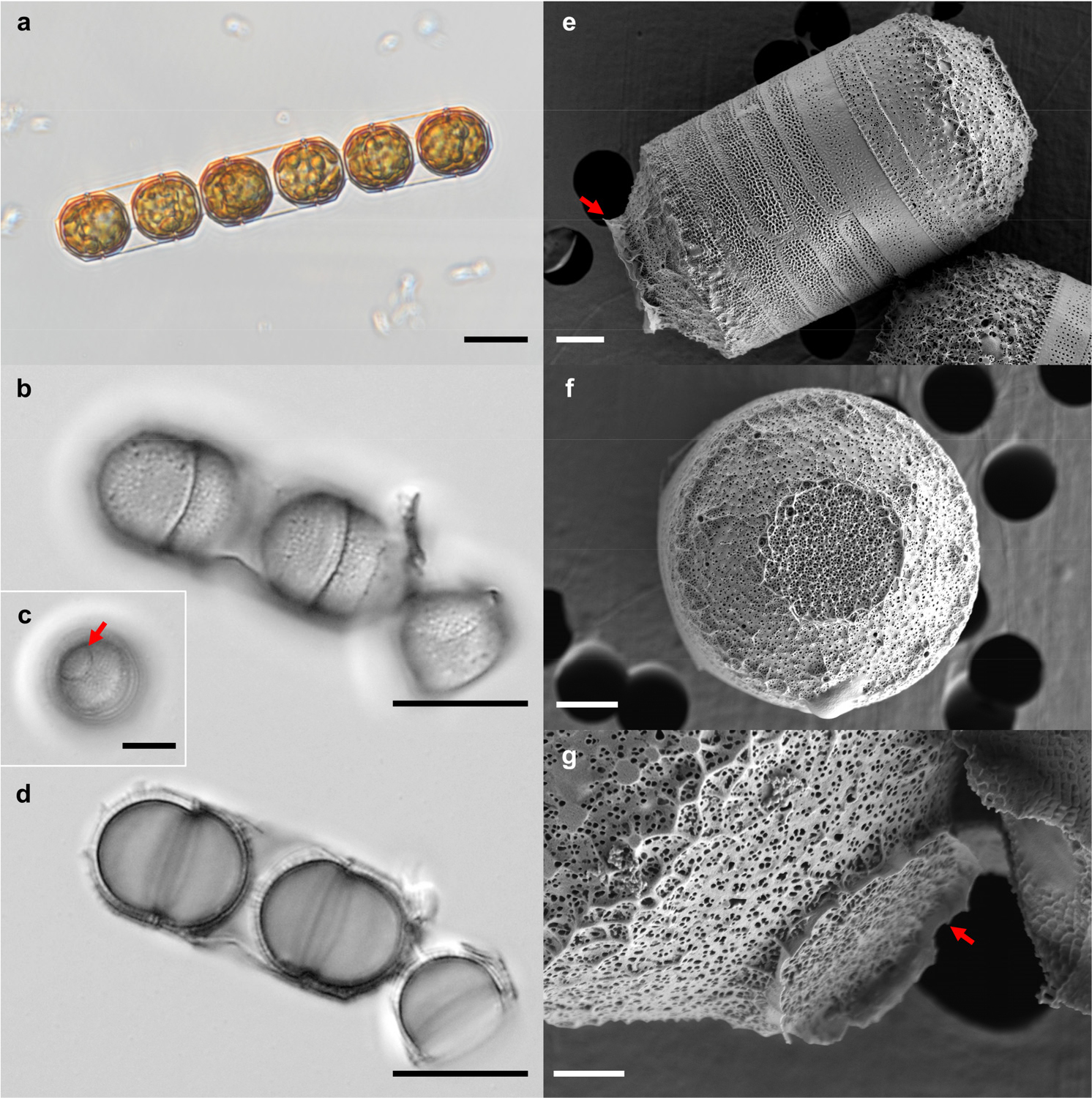
Fig. 2.
Light and scanning electron micrographs of M. octogona MOJJ22. (a) Light micrograph of the girdle view of six live cells forming a chain-type colony. (b–d) Light micrograph of cleaned frustules. (b) and (d) represent same frustules at different focus levels. (c) showing the corona (arrow) on valve face. (e) The whole frustule in the girdle view showing the bands of the cingulum. The well-developed corona is shown by the arrow. (f) Exterior of the valve surface. (g) Details of the frustule, indicating the corona (arrow). Scale bars: a, b & d = 10 µm, c = 5 µm, e & f = 2 µm, g = 1 µm
The valve by SEM analysis exhibited a hemispherical shape but sloped upwards to the valve mantle at an angle of approximately 40° from approximately half of the center of the valve (Fig. 2e). The plain flange-shaped corona was well-developed in the middle of the valve face (Fig. 2e and g). The valve interior within the corona exhibited a randomly arranged irregular pattern of polygonal loculae containing simple pores (Fig. 2f). The slope of the valve face exhibited an irregular pattern of distinct stellate thickening with simple pores (Fig. 3a). The rimoportulae, situated on the valve face within the corona and valve mantle, exhibited a rounded to elliptical shape, and were externally rimmed (Fig. 2b). The rimoportulae on the inclined portion of the valve face exhibited pronounced stellate thickening (Fig. 3a and b). The rimmed mantle rimoportulae were arranged in a marginal ring near the mantle edge (Fig. 3c), with 4–5 within 10 μm. The mantle was milled at its edges (Fig. 3c). Internally, the rimoportulae exhibited a consistent shape, irrespective of their external morphology, and were observable as small, elongated fissures (Fig. 3d). The loculate areolae were internally closed by individual vela (Fig. 3d). The entire epicingulum consisted of six open cingular bands, all of which were ligulate (Fig. 3e). The ligulae exhibited a long, narrow morphology (Fig. 3c and e). In contrast to the valve face, each band displayed perforations in the form of straight, longitudinal rows of circular to elongated pores (60–65 rows within 10 μm) with hyaline strips surrounding the margin (Fig. 3c and e). Valvocopula exhibited a lower pore density (45–50) than the other bands (Fig. 3c, and e).
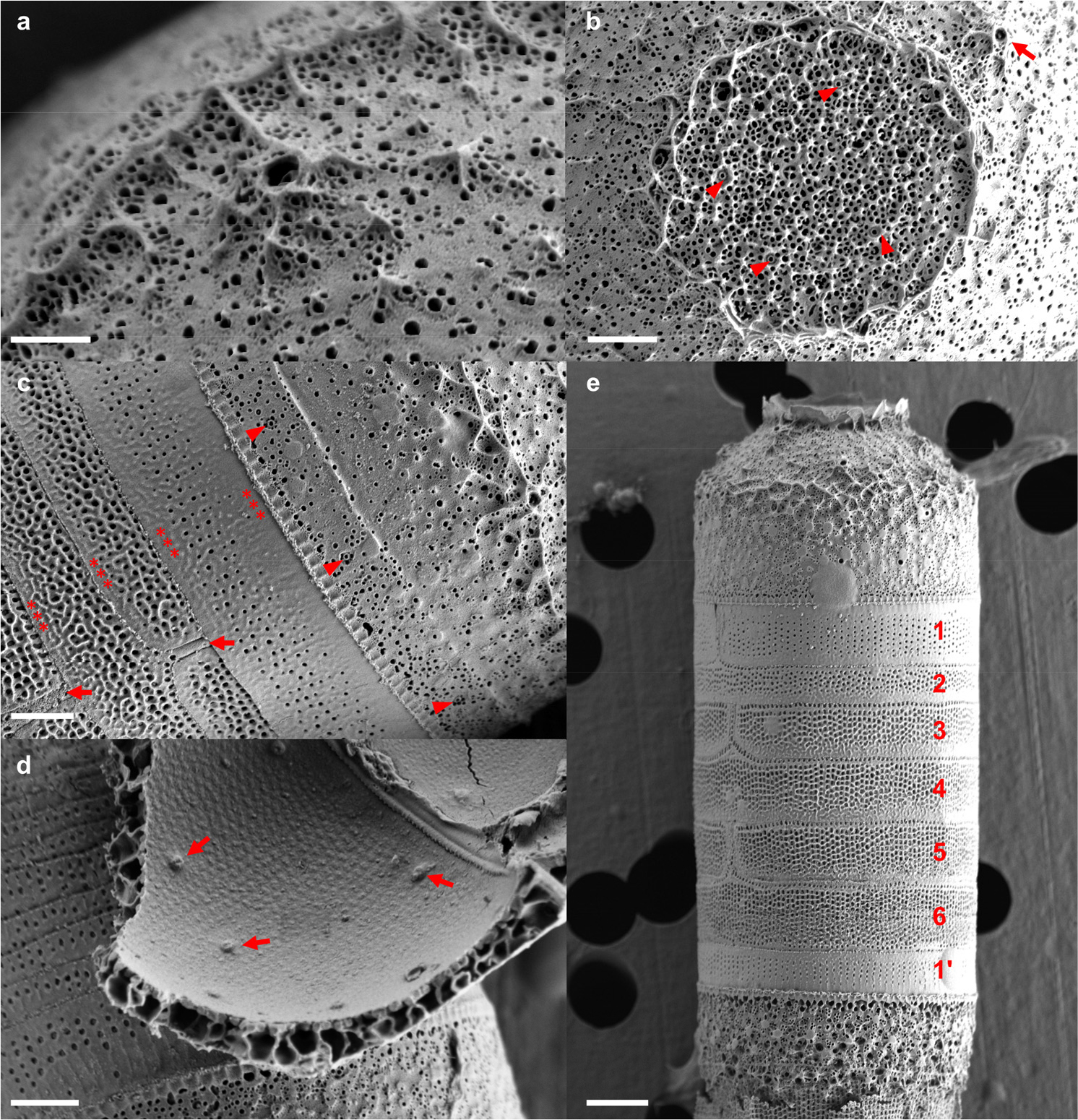
Fig. 3.
Scanning electron micrographs of M. octogona MOJJ22. (a) Details of the rimoportula located inclined of the valve face. A strongly stellate thickening was present around the rimoportula. (b) External valve face, detail of the center. The rimmed rimoportulae in the valve center are encircled by well-developed coronas (arrowheads). (c) The junction of valve mantle with valvocopula. The ring of mantle rimoportulae (arrowheads) milled the mantle edge nearby. The cingulum is also shown; the hyaline strip (asterisks) and ligulae (arrows) of bands are shown. (d) The interior of the broken valve showing irregularly located rimoportulae (arrows). A chambered (loculate) valve is shown. (e) General view of cingulum in a large frustule. The complete girdle, composed of the epicingulum of bands 1–6 and hypocingulum of which only band 1' is visible, is shown. Scale bars: a = 0.5 µm, b–d = 1 µm, e = 2 µm
M. octogona can be easily confused with M. moniliformis var. octogona (Grunow) Hustedt 1927 (fig. 1 pl. 7-9). In general, live cells of Melosira species are predominantly observed in the girdle view, as these two species possess an octagonal shape in the girdle view. Furthermore, the challenge lies in its limited ability to differentiate owing to the lack of a comprehensive understanding of the morphological characteristics of M. octogona. The most significant distinction between the two species is the presence or absence of a corona on the valve face. Unlike M. octogona (Fig. 2c and g), M. moniliformis var. octogona can be readily distinguished by the absence of a corona on the valve surface, but rather by the presence of small spines arranged in an irregular pattern (Crawford 1977; Yamamoto 2019: fig. 2-2e and f). Another distinctive characteristic of M. octogona is the presence of stellate thickening on its valve surface (Figs. 2f and 3a), which distinguishes it from other Melosira species. The morphology of these valve surfaces was somewhat similar to that observed in the resting spore of Melosira arctica Dickie (Fernandes and Souza-Mosimann 2001; Kaczmarska and Jahn 2006). Additionally, the valve diameter of M. octogona seems to be smaller (10.4 to 11.1 μm) compared to that of M. moniliformis var. octogona (15 to 34 μm) (Edlund et al. 2001; Al-Yamani and Saburova 2011; Yamamoto 2019).
Phylogenetic analysis
In this study, partial gene sequences of the 18s rDNA (1,665 bp) of Melosira octogona were acquired for the first time. The sequence was deposited in GenBank under accession number PP263059. Phylogenetic analysis was performed to determine the relationship between M. octogona and the 23 species of the order Melosirales obtained from GenBank. Both ML and BI methods yielded similar topologies in the phylogenetic trees (Fig. 4). Phylogenetic analysis strongly supported M. octogona as a sister to Melosira sp. strain 52 (KJ961704), with a bootstrap value of 98 and a posterior probability of 100. The 18S rDNA sequence of our strain exhibited 100% similarity with this sequence, with a query coverage of 100% and an E-value of 0. Among the sequences identified at the species level, more than 99.46% similarity was observed (with a query cover of 100% and an E value of 0) with M. dubia Kützing (Synonym of Podosira dubia (Kützing) Grunow) (AB430588). Furthermore, it exhibited 99.03% similarity with Melosira cf. octogona (AY485518). Nevertheless, the number of SSU sequences from Melosira species documented in GenBank is limited, and the majority of the highly ranked sequences in the BLAST analysis are unpublished, making it challenging to attribute substantial significance to them.
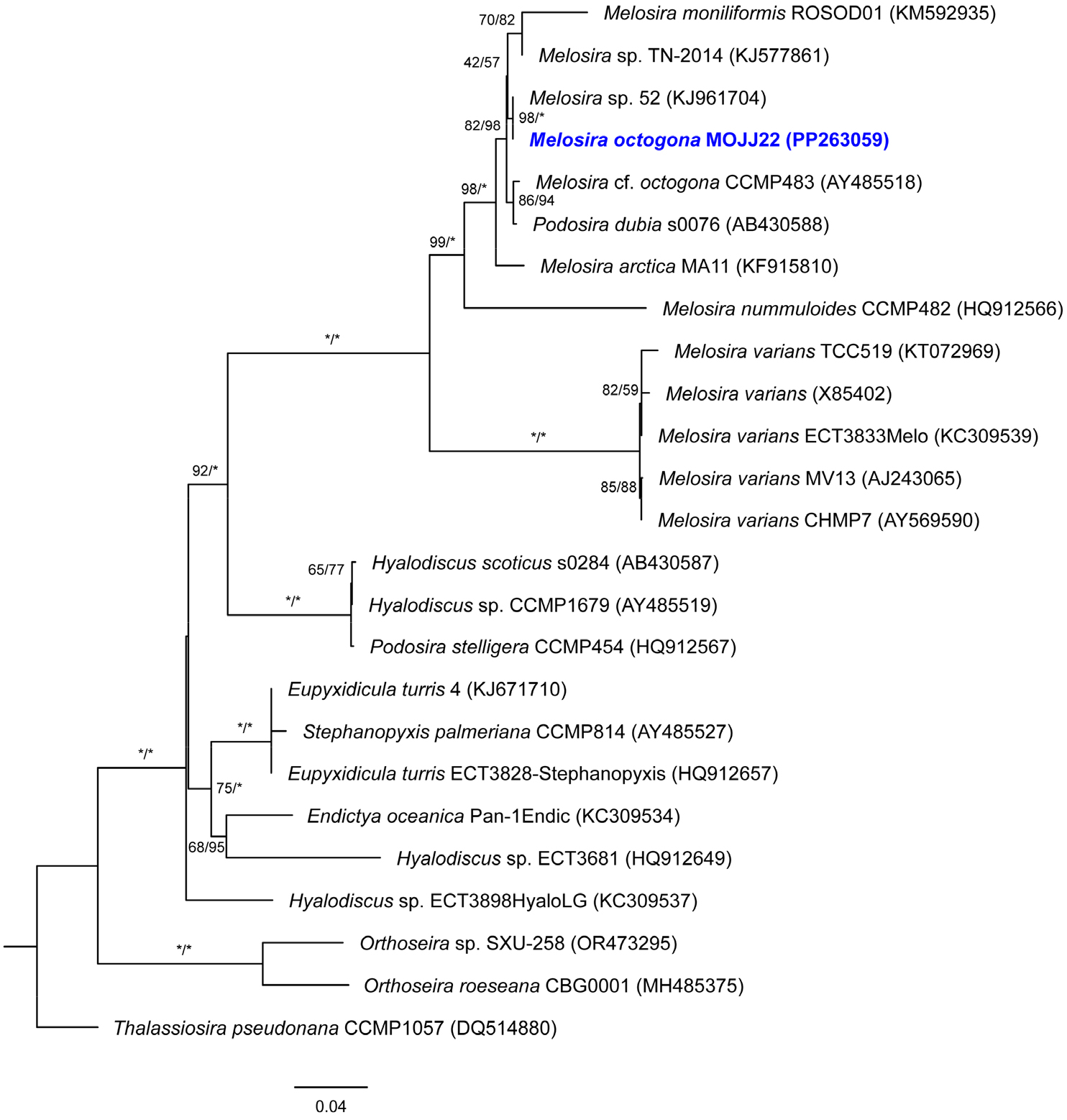
Fig. 4.
ML and BI phylogenetic trees of 18S rDNA from Melosirales species. The values for each node indicate the ML bootstrap and Bayesian posterior probabilities (%), respectively. The asterisk (*) indicates 100. Thalassiosira pseudonana was used as the outgroup. M. octogona MOJJ22 is highlighted in bold blue
In conclusion, this study examined the morphological characteristics of Melosira octogona using SEM, and analyzed the small subunit rDNA sequence of the strain retrieved from the coast of Jeju Island, South Korea. The results of our analysis can enhance the precise recognition of M. octogona and enrich our understanding of its phylogenetic position within the order Melosirales. However, further research is warranted to acquire comprehensive taxonomic information for a better understanding of the classification and phylogeny of Melosira species.





