1. Introduction
Sculpins as common benthic fish are distributed in most oceans of the world (Nelson 2006; Johannesen et al. 2012). The Cottidae is the largest family of sculpins, which comprise more than 70 genera and 275 species worldwide (Nelson 2006). They mainly occur in boreal and cold-temperate regions across the Arctic (Nelson 2006). Although sculpins are not normally deemed a suitable food source for higher tropic levels (e.g., fish, seabirds, and mammals) in the Arctic, they probably form an important resource contributing to food web stability, considering their abundance and availability in dynamically-changing environments in the Arctic (e.g., seasonal ice cover and open water) (Rooney et al. 2006; Walsh 2008). The genus Triglopsis has a wide distribution in the northern hemisphere with noticeable variations in morphology. Fourhorn sculpin Triglopsis quadricornis Linnaeus, 1758 is observed in nearshore open water in the Arctic (COSEWIC 2003) However, little is known about their general biology (e.g., reproduction, feeding strategies), habitats, population sizes or trophic roles.
In this study, we present the complete mitogenome of T. quadricornis (Accession no. MT789711; registered with homotypic synonym Myoxocephalus quadricornis) by de novo assembling from Illumina shotgun sequence data. An individual of T. quadricornis was sampled at J. P. Koch Fjord near Sirius Passet, North Greenland (82°46' 1.7"N, 42°32'52.4"W; Lee 2018) on 16th July 2017. The voucher specimen was registered in the Korea Polar Research Institute (KOPRI; Species ID: SP-fish; Specimen ID: 170716_ SP-fish01). Genomic DNA was isolated from muscle tissue by using a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). Quantity and quality of the extracted genomic DNA were measured using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). A genomic library was constructed using TruSeq Nano DNA Kit by a commercial company (Macrogen, Seoul, South Korea) according to the manufacturer's instructions (Illumina, San Diego, CA, USA). The sequencing library was constructed by random fragmentation of the genomic DNA with 5’ and 3’ adapter ligation. The ligation product was amplified by PCR before loading into a flow cell. Raw reads were obtained from the sample that passed quality control by Illumina HiSeq platform. Trimming and filtering were performed using FastP and Trimmomatics, and 24,643,780 reads were finally obtained. After the trimming process, de novo assembly was performed with SPAdes v3.11.1 (Bankevich et al. 2012). Genomic features and annotations were performed using MITOS2 (Bernt et al. 2013) and tRNAscan-SE 2.0 (Lowe and Eddy 1997). The annotated gene structure was further confirmed using NCBI- BLAST (http://blast.ncbi.nlm.nih.gov).
The mitochondrial genome content of T. quadricornis was found to generally include a typical set of 13 PCGs, 22 tRNAs, and the large and small rRNA of the ribosome as observed in Perciformes species (Mascolo et al. 2019; Ceruso et al. 2020). The full circular contig was 16,736 bp in length with a GC content of 46.7% and an AT bias (A: 26.8%; T: 26.4%; G: 17.4%; C: 29.3%). Recently, the complete mitochondrial genomes of two individuals of M. quadricornis (Accession no. MT303953 and MT303954) collected from the White-Sea were released (Balakirev et al. 2020b). Although the two M. quadricornis mitochondrions have a length of 16,682 bp, identity between the individuals was 99.8% (37/16,682 bp). When we compared T. quadricornis mitochondrion to the two mitogenomes of M. quadricornis, slight differences (0.4~0.9%) in similarity were observed in ND1, ND2, ND5, and CYTB genes. Overall, gene composition, arrangement, and other features (e.g., size, GC ratio) are highly similar to the mitogenomes registered in the genus Myoxocephalus and Megalocottus (Balakirev et al. 2020a; Li et al. 2018). In the T. quadricornis mitogenome, COI gene has GTG as a start codon and three genes, COII, ND4, and CYTB have an incomplete stop codon (T--), as shown in the genus Myoxocephalus (Li et al. 2018).
A maximum likelihood phylogenetic hypothesis was established using sequence data from the concatenated set of 13 PCGs of T. quadricornis mitogenome with including 21 published mitogenomes from the Cottidae. JModelTest ver. 2.1.10 (Darriba et al. 2012) was used to select the best substitution model and a substitution model (HKY + G + I) was applied to perform a maximum-likelihood (ML) method in the PhyML 2.4.5 (Guindon and Gascuel 2003) with 1000 bootstrap replicates. The mitogenome of T. quadricornis in this study is clustered together with Megalocottus and Myoxocephalus species (Fig. 1). In addition, our COI sequence was identical to the partial COI sequences of T. quadricornis (652 bp; GenBank accession no. EU524913) and M. quadricornis (648 bp; GenBank accession no. MG421040), whereas similarity between T. quadricornis in this study and Megalocottus platycephalus (1,009 bp; GenBank accession no. MH016197) was 96%. Complete mitochondrial genomes have been recently released from two local specimens of Myoxocephalus quadricornis (probably synonym of Triglopsis quadricornis) (MQI1, GenBank accession no. MT303953.1 and MQI3, MT303954.1), which were collected from Dvina Bay, White Sea, Russia. Difference of the whole mitochondrial DNA sequence between MQI1 and MQI3 turns out to be only 0.2%, while those of the both White Sea specimens differ from the Greenland specimen in this study appears by 0.6%. The differences in mitochondrial DNA sequence, therefore, would have been derived by different geographical distributions, but additional mitogenome data are required to understand the differences in mitogenomes in this genus.
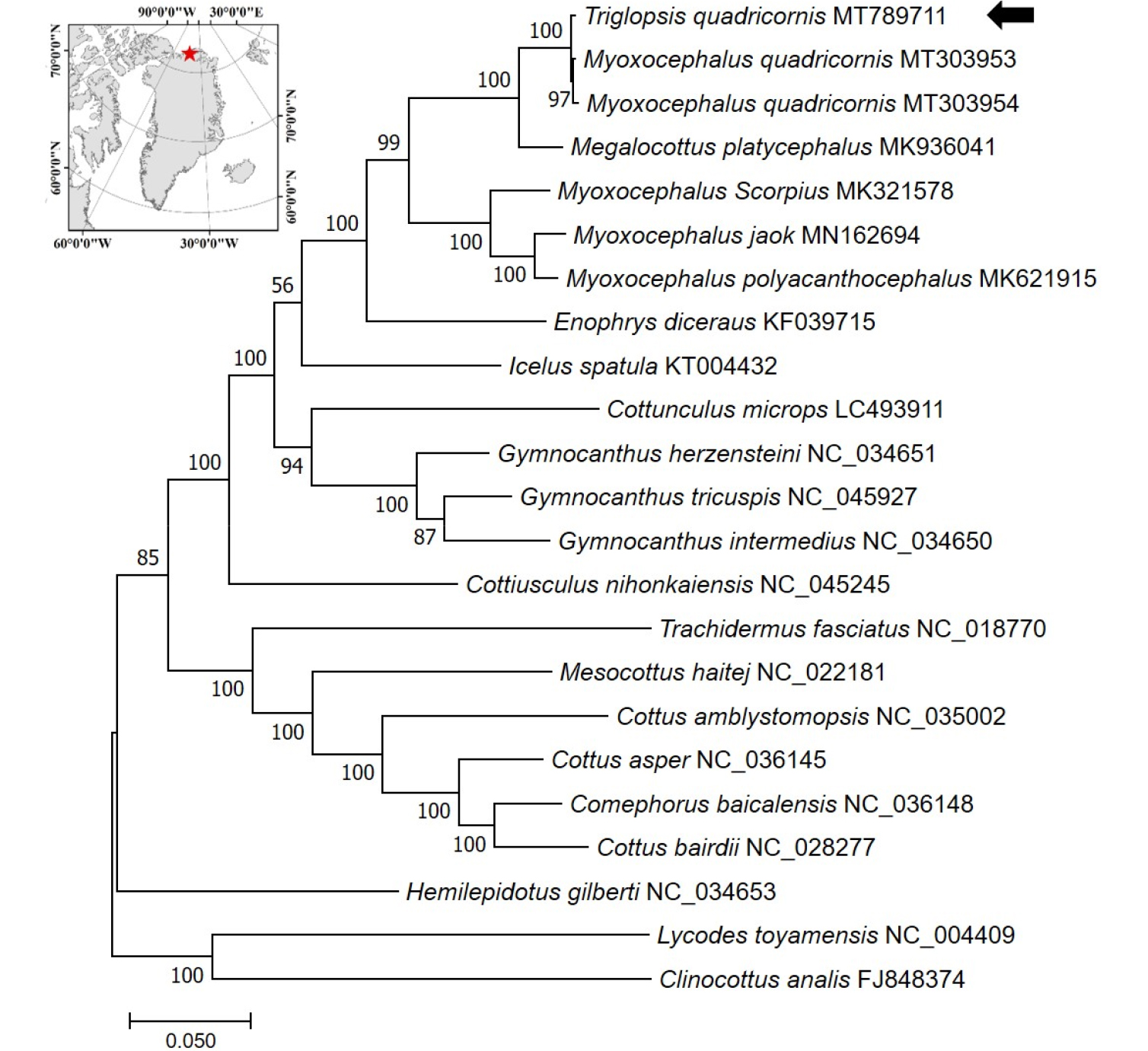
Fig. 1.
Maximum-likelihood (ML) phylogeny of 20 species of the family Cottidae based on the concatenated nucleotide sequences of entire protein-coding genes (PCGs). Numbers on the branches indicate ML bootstrap percentages (1000 replicates). DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated. The black arrow indicates the T. quadricornis analyzed in this study
2. Data Availability Statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/ MT789711. Associated BioProject, SRA, and Bio Sample accession numbers are https://www.ncbi.nlm.nih.gov/biopro ject/PRJNA673747, https://www.ncbi.nlm.nih.gov/sra/SRR12 964012, and https://www.ncbi.nlm.nih.gov/biosample/SAMN 16622464, respectively.




